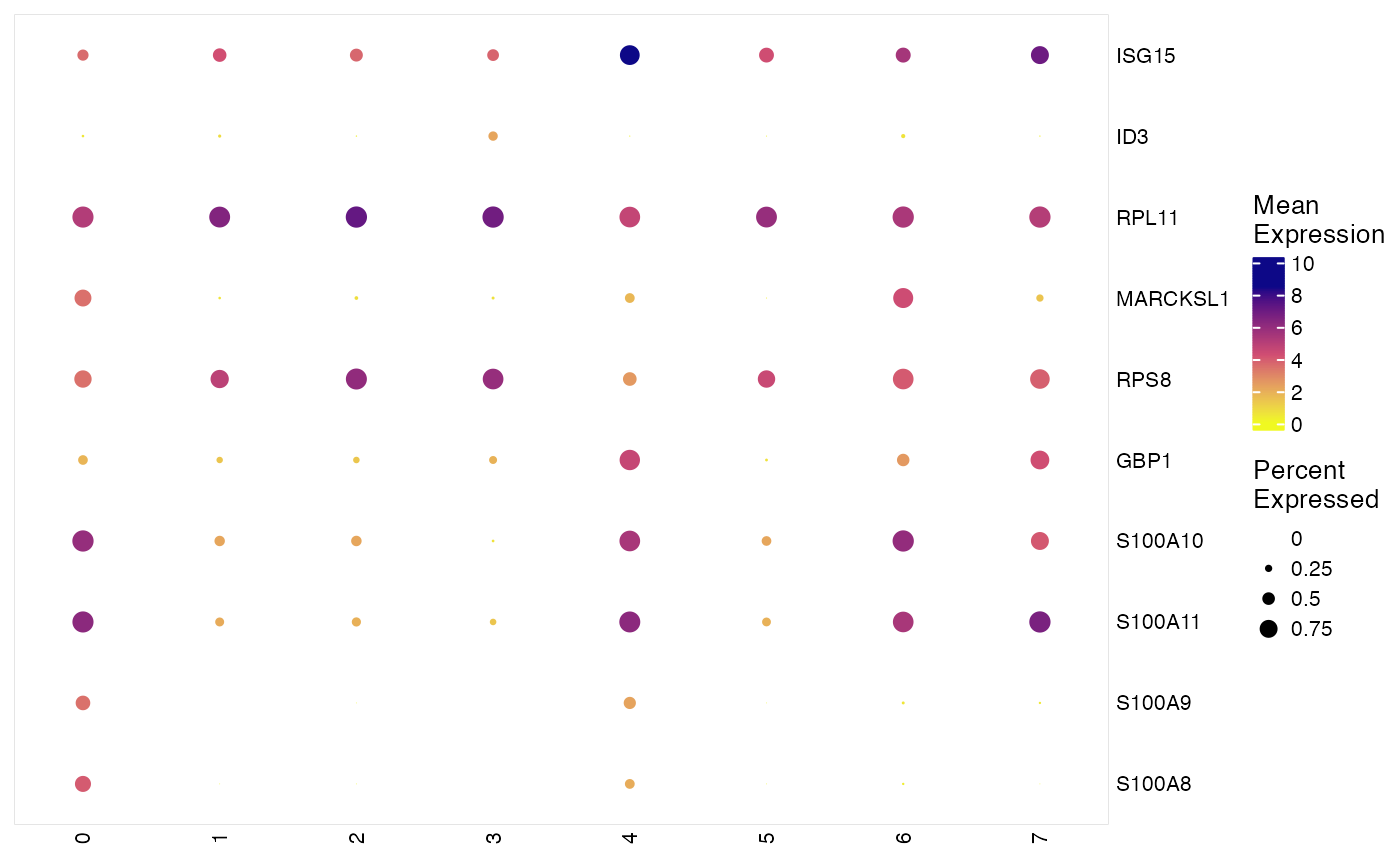
This function produces dot plots. Each column represent a group
of cells specified by groupBy, each row is a gene specified by
features. The color of dots reflects mean of normalized expression of
specified genes in each cell group and sizes reflects the percentage of cells
expressing each gene in a group. We utilize
ComplexHeatmap
for simplified management of adding annotation and slicing subplots. This was
inspired by the implementation in
scCustomize.
Usage
plotClusterGeneDot(
object,
features,
groupBy = NULL,
splitBy = NULL,
featureScaleFunc = function(x) log2(10000 * x + 1),
cellIdx = NULL,
legendColorTitle = "Mean\nExpression",
legendSizeTitle = "Percent\nExpressed",
viridisOption = "magma",
verbose = FALSE,
...
)Arguments
- object
A liger object
- features
Use a character vector of gene names to make plain dot plot like a heatmap. Use a data.frame where the first column is gene names and second column is a grouping variable (e.g. subset
runMarkerDEGoutput)- groupBy
The names of the columns in
cellMetaslot storing categorical variables. Expression data would be aggregated basing on these, together withsplitBy. Default uses default clusters.- splitBy
The names of the columns in
cellMetaslot storing categorical variables. Dotplot panel splitting would be based on these. DefaultNULL.- featureScaleFunc
A function object applied to normalized data for scaling the value for better visualization. Default
function(x) log2(10000*x + 1)- cellIdx
Valid cell subscription. See
subsetLiger. DefaultNULLfor using all cells.- legendColorTitle
Title for colorbar legend. Default
"Mean\nExpression".- legendSizeTitle
Title for size legend. Default
"Percent\nExpressed"- viridisOption
Name of available viridis palette. See
viridis. Default"magma".- verbose
Logical. Whether to show progress information. Mainly when subsetting data. Default
FALSE.- ...
Additional theme setting arguments passed to
.complexHeatmapDotPlotand heatmap setting arguments passed toHeatmap. See Details.
Value
HeatmapList object.
Details
For ..., please notice that arguments colorMat,
sizeMat, featureAnnDF, cellSplitVar, cellLabels
and viridisOption from .complexHeatmapDotPlot are
already occupied by this function internally. A lot of arguments from
Heatmap have also been occupied: matrix,
name, heatmap_legend_param, rect_gp, col, layer_fun, km, border, border_gp,
column_gap, row_gap, cluster_row_slices, cluster_rows, row_title_gp,
row_names_gp, row_split, row_labels, cluster_column_slices, cluster_columns,
column_split, column_title_gp, column_title, column_labels, column_names_gp,
top_annotation.
Examples
# Use character vector of genes
features <- varFeatures(pbmcPlot)[1:10]
plotClusterGeneDot(pbmcPlot, features = features)
 # Use data.frame with grouping information, with more tweak on plot
features <- data.frame(features, rep(letters[1:5], 2))
plotClusterGeneDot(pbmcPlot, features = features,
clusterFeature = TRUE, clusterCell = TRUE, maxDotSize = 6)
# Use data.frame with grouping information, with more tweak on plot
features <- data.frame(features, rep(letters[1:5], 2))
plotClusterGeneDot(pbmcPlot, features = features,
clusterFeature = TRUE, clusterCell = TRUE, maxDotSize = 6)
