Welcome to the Welch Lab
Our research aims to address fundamental problems in both biomedical research and computer science by developing new tools tailored to rapidly emerging high-throughput sequencing technologies. Broadly, we seek to understand what genes define the complement of cell types and cell states within healthy tissue, how cells differentiate to their final fates, and how dysregulation of genes within specific cell types contributes to human disease. As computational method developers, we seek to both employ and advance the methods of machine learning, particularly for unsupervised analysis of high-dimensional data.
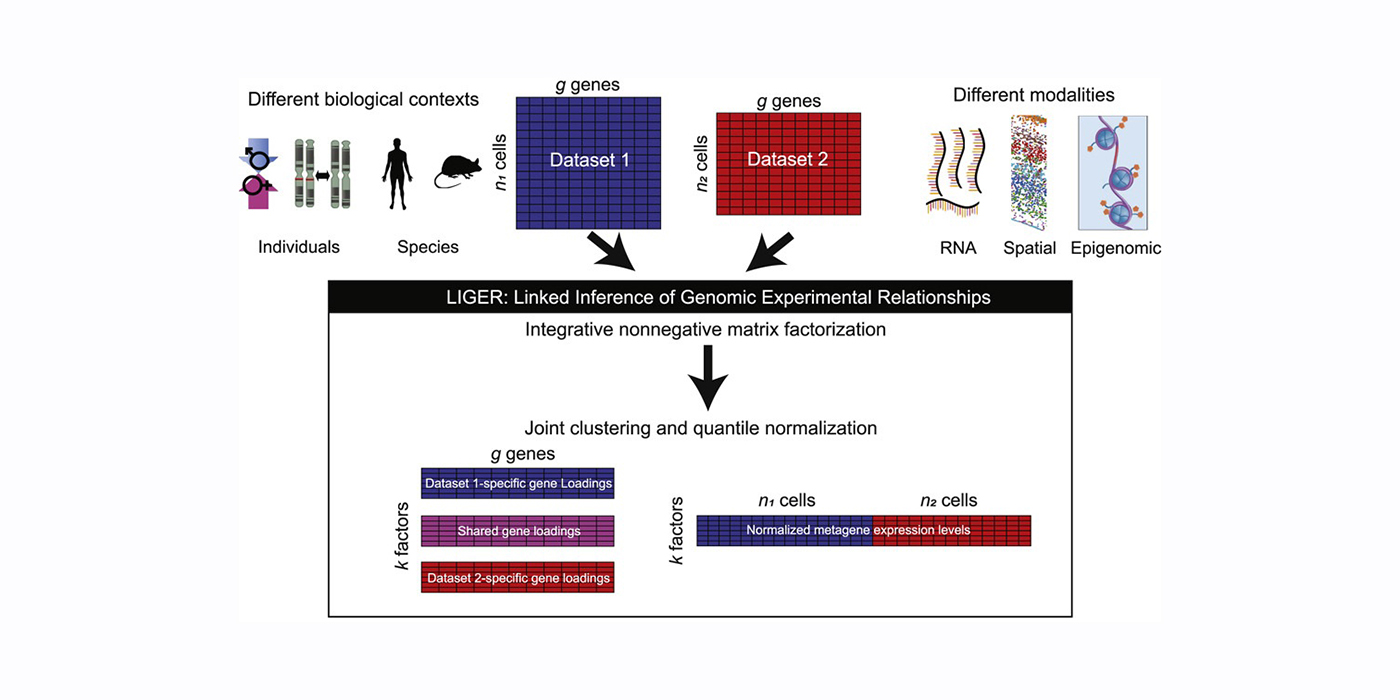
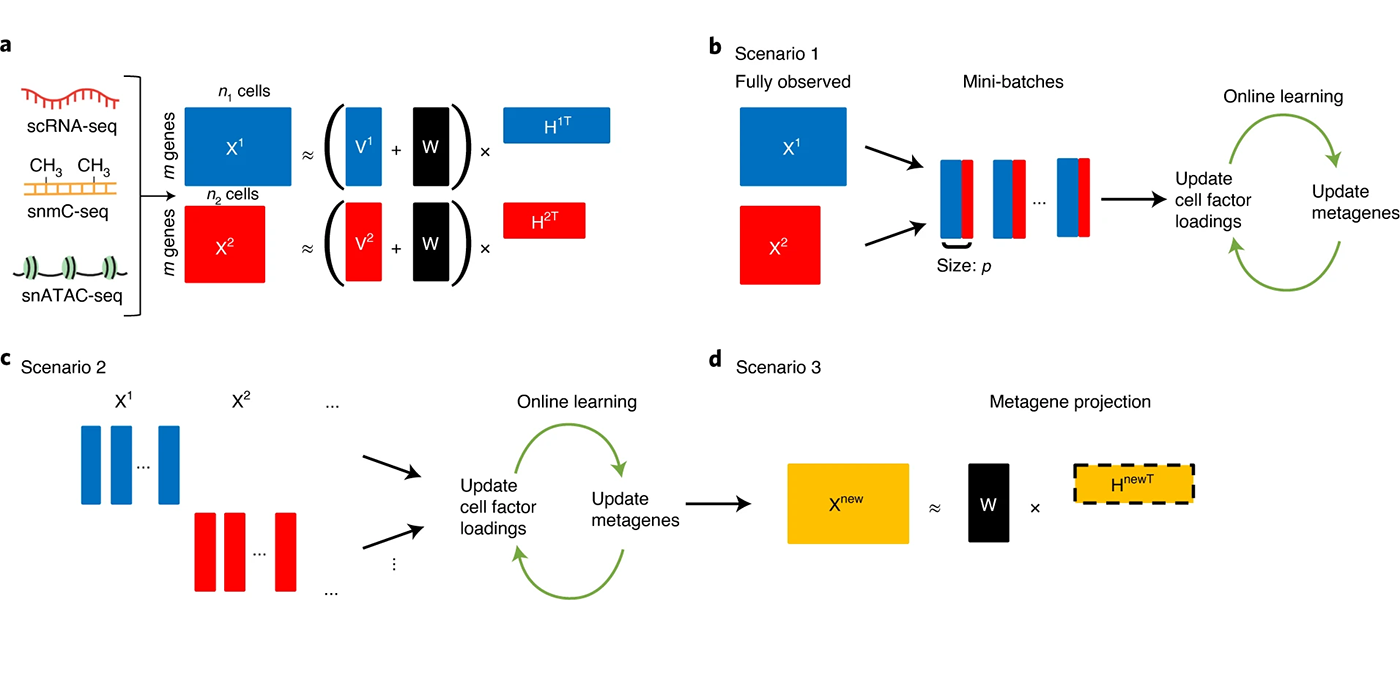
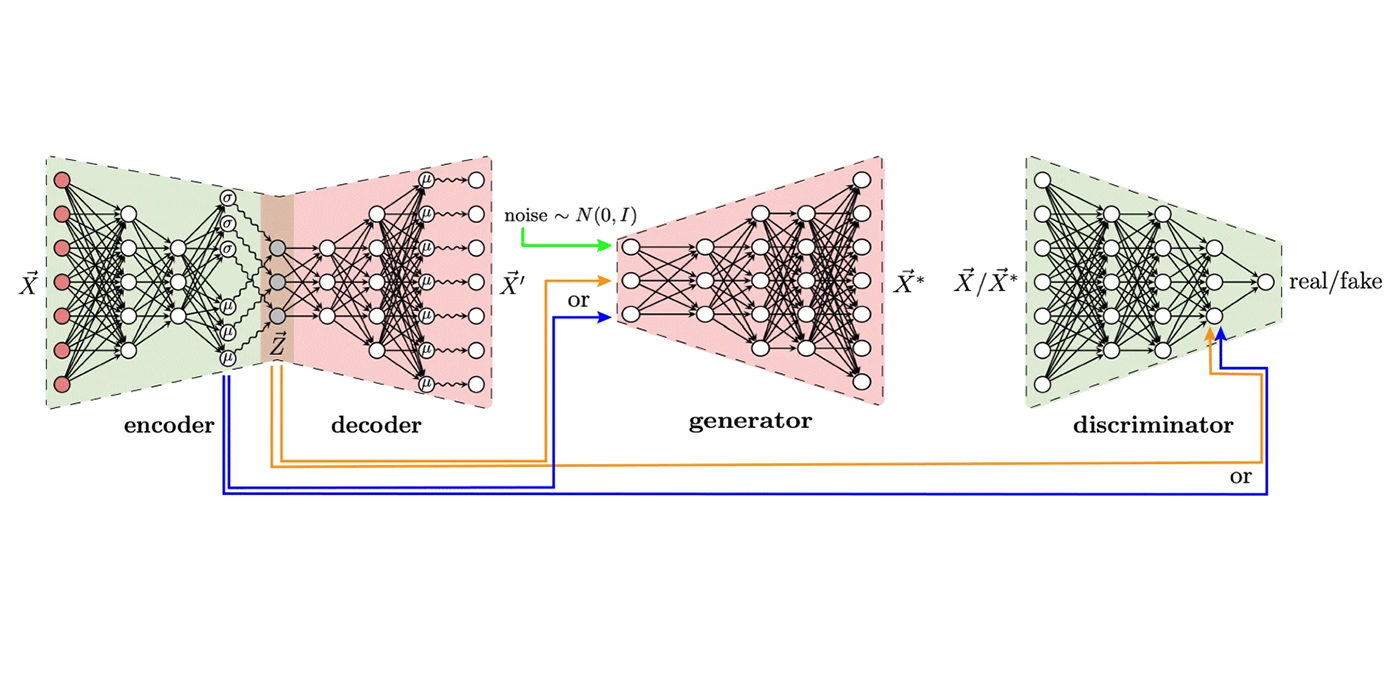
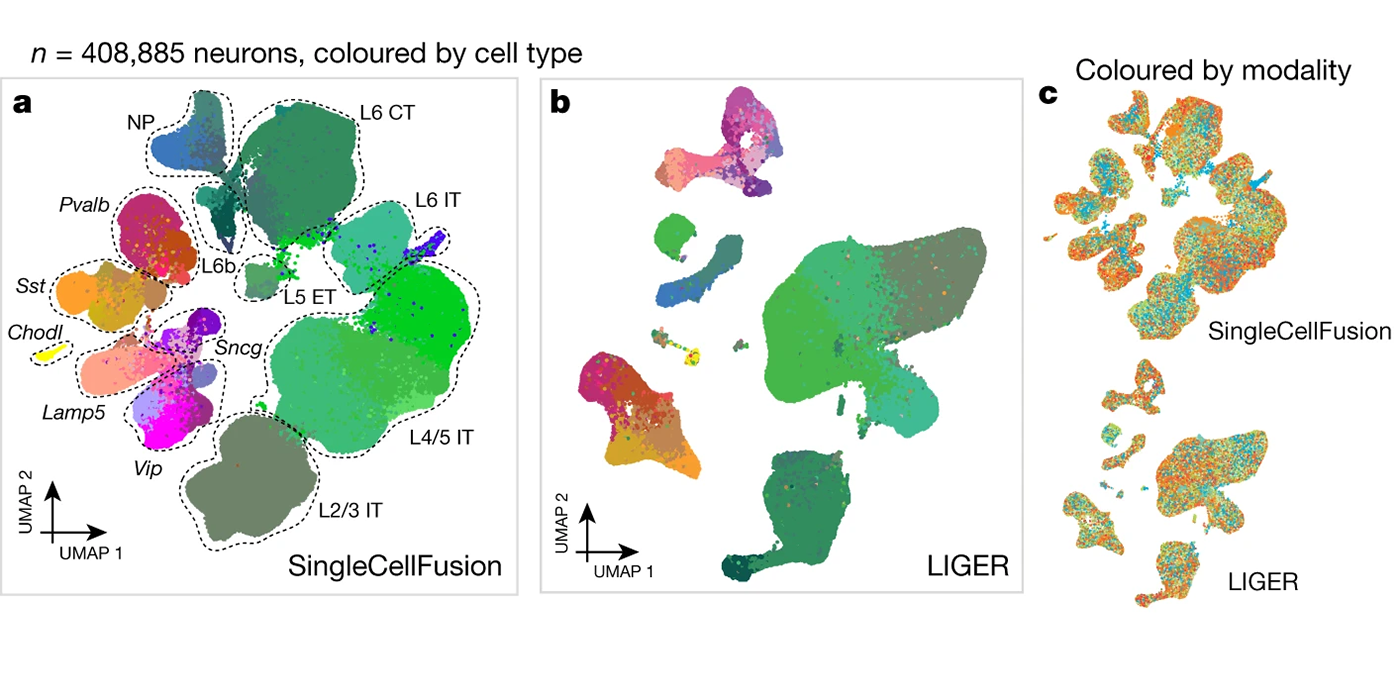
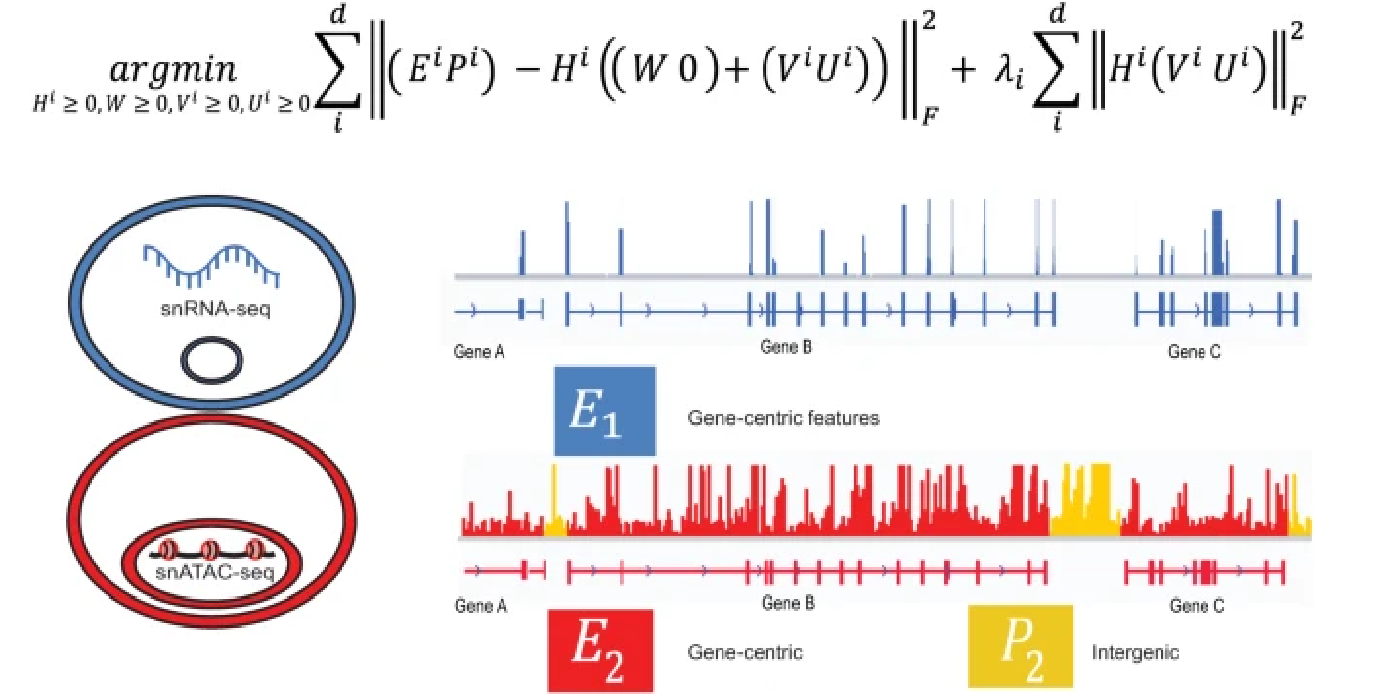
Most recently, I have focused on developing open-source software for the processing, analysis, and modeling of single-cell sequencing data. Key contributions in this area include LIGER, a general approach for integrating single-cell transcriptomic, epigenomic and spatial transcriptomic data; online iNMF, a scalable and iterative algorithm for single-cell data integration; and MultiVelo, a tool for modeling cell fate transitions from single-cell multi-omic data. I have applied these methods in collaboration with biological scientists to study stem cell differentiation, somatic cell reprogramming, and the mammalian brain.
The Welch Lab has openings for multiple positions, including Postdoctoral Fellow, PhD Student, Bioinformatician, and Software Engineer! (more info)
News
November 26, 2025Crystal's paper is published in Genome Biology
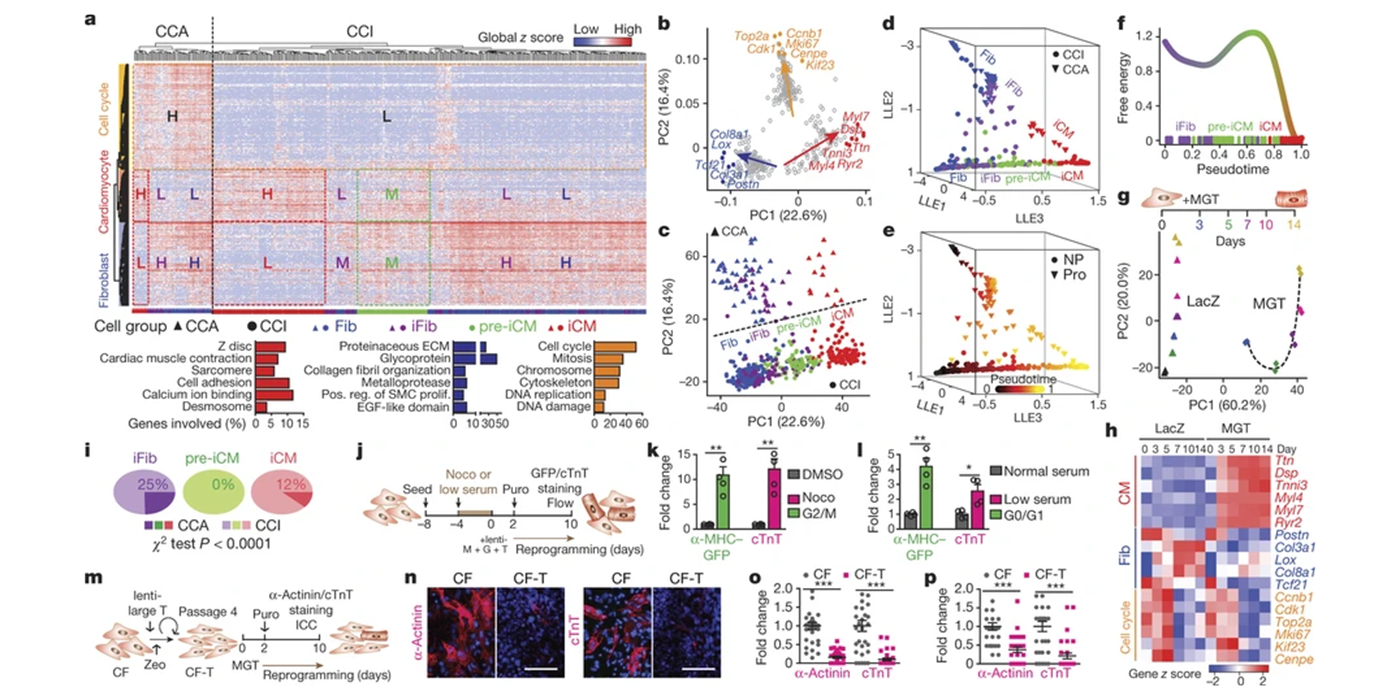
November 20, 2025Chen's paper is published in Nature Communications
September 17, 2025Yuxuan's paper is published in Genome Biology
September 1, 2025NIH/NIDA multi-PI R33 awarded to Collins and Welch labs
August 4, 2025PerturbNet paper featured as cover article
July 17, 2025UM press release highlights TopoVelo paper
July 16, 2025Research briefing for TopoVelo paper is published in Nature Biotechnology
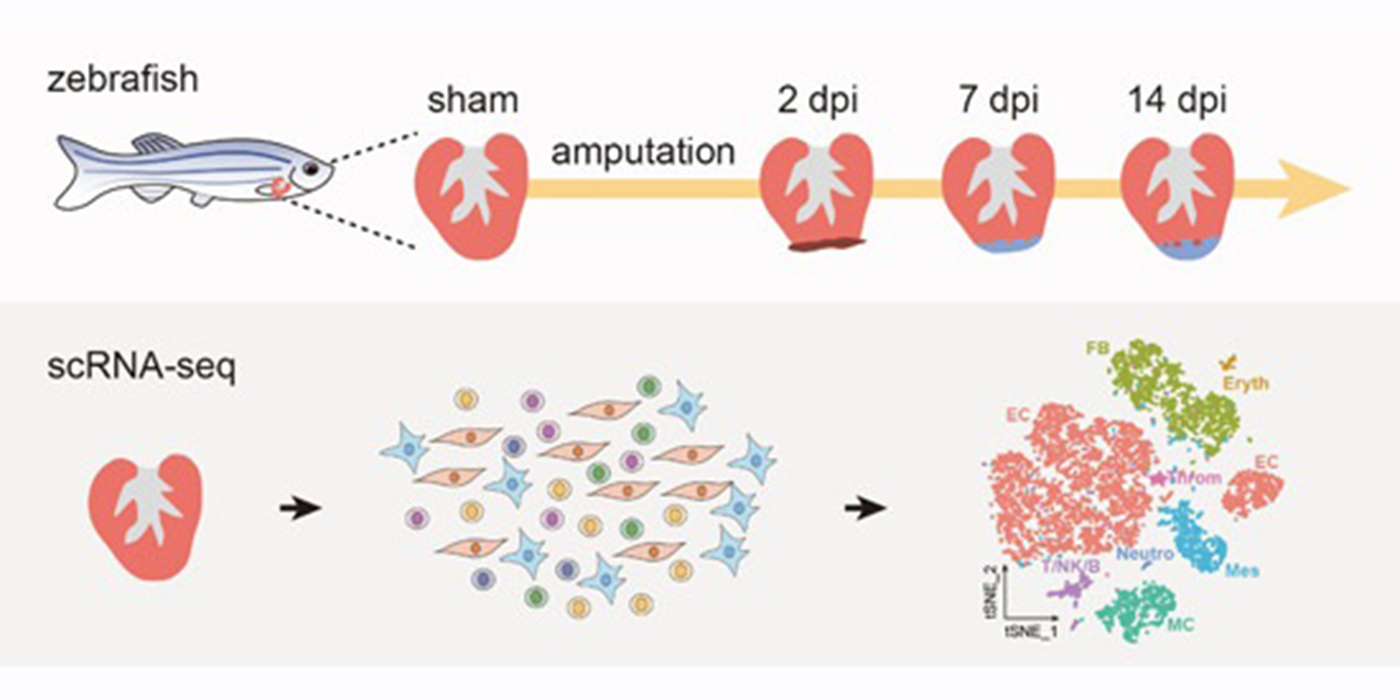
July 16, 2025Yichen, Jialin, and Justin's paper is published in Nature Biotechnology
July 10, 2025Hengshi and Weizhou's paper is published in Molecular Systems Biology