Publications
Group highlights
(For a full list of publications see below or go to PubMed, Google Scholar, or Michigan Experts)
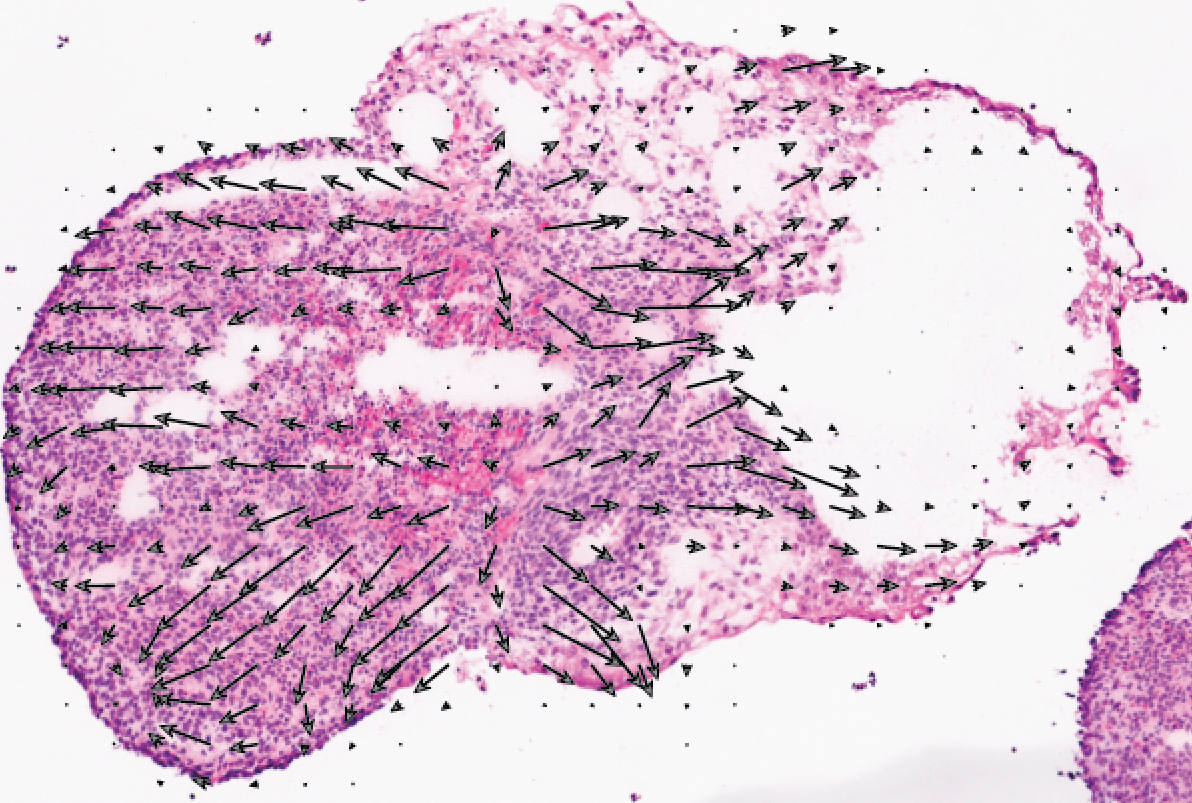
TopoVelo extends the RNA velocity framework to model single-cell gene expression dynamics of an entire tissue with spatially coupled differential equations. TopoVelo infers the direction of cell migration and differentiation, the degree of spatial cell influence, and morphological changes. We generate Slide-seq data from an in vitro model of human development and use TopoVelo to study the spatial patterns of early differentiation.
Gu Y*, Liu J*, Lee KH*, Li C, Lu L, Moline J, Guan R, Welch JD
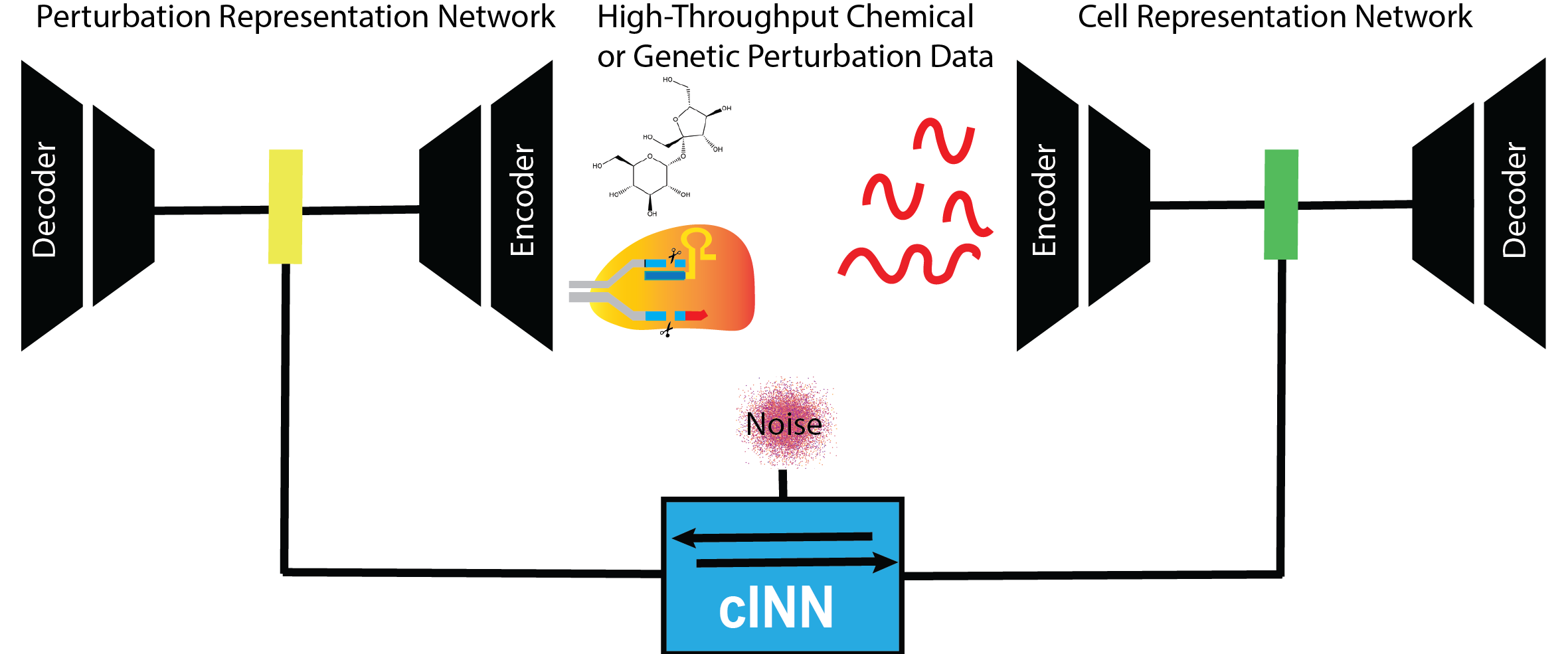
PerturbNet is a generative AI model that can predict shifts in cell state—changes in overall gene expression—in response to multiple types of unseen cellular perturbations. PerturbNet uses a flexible framework to “mix and match” neural networks and predict effects of chemical, gene knockdown, gene overexpression, and DNA sequence mutations. Despite being an all-in-one model, PerturbNet shows competitive performance with previous methods tailored to predict either chemical or genetic effects. An in silico screen of all possible single amino acid substitutions in GATA1 identifies candidates likely to modify erythroid cell differentiation.
Yu H*, Qian W*, Song Y, Welch JD
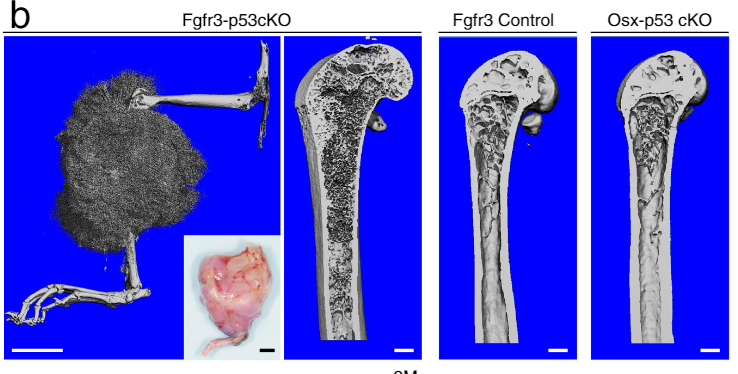
Using scRNA and snATAC analysis, we identified a new class of skeletal stem cells called osteoblast-chondrocyte transitional stem cells. OCT stem cells give rise to three types of mature bone cells after bone injury, are depleted in old mice, and cause aggressive osteosarcomas upon conditional p53 knockout.
Matsushita Y*, Liu J*, Ka Yan Chu A, Tsutsumi-Arai C, Nagata M, Arai Y, Ono W, Yamamoto K, Saunders TL, Welch JD*, Ono N*
*Co-First Authors, Co-Senior Authors
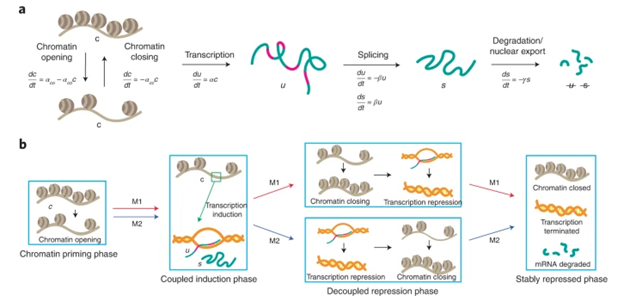
Cells differentiate to their final fates through sequential epigenetic and transcriptional changes. A mathematical model fit on multi-omic single-cell data yields insights into the temporal relationships between chromatin accessibility and gene expression during cell differentiation.
Li C, Virgilio MC, Collins KL, Welch JD
Also accepted at RECOMB 2022.
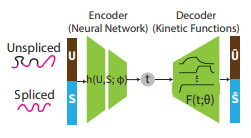
By using a simple family of ODEs informed by the biochemistry of gene expression to constrain the likelihood of a deep generative model, we can simultaneously infer the latent time and latent state of each cell and predict its future gene expression state. The model can be interpreted as a mixture of ODEs whose parameters vary continuously across a latent space of cell states.
Gu Y, Blaauw D, Welch JD
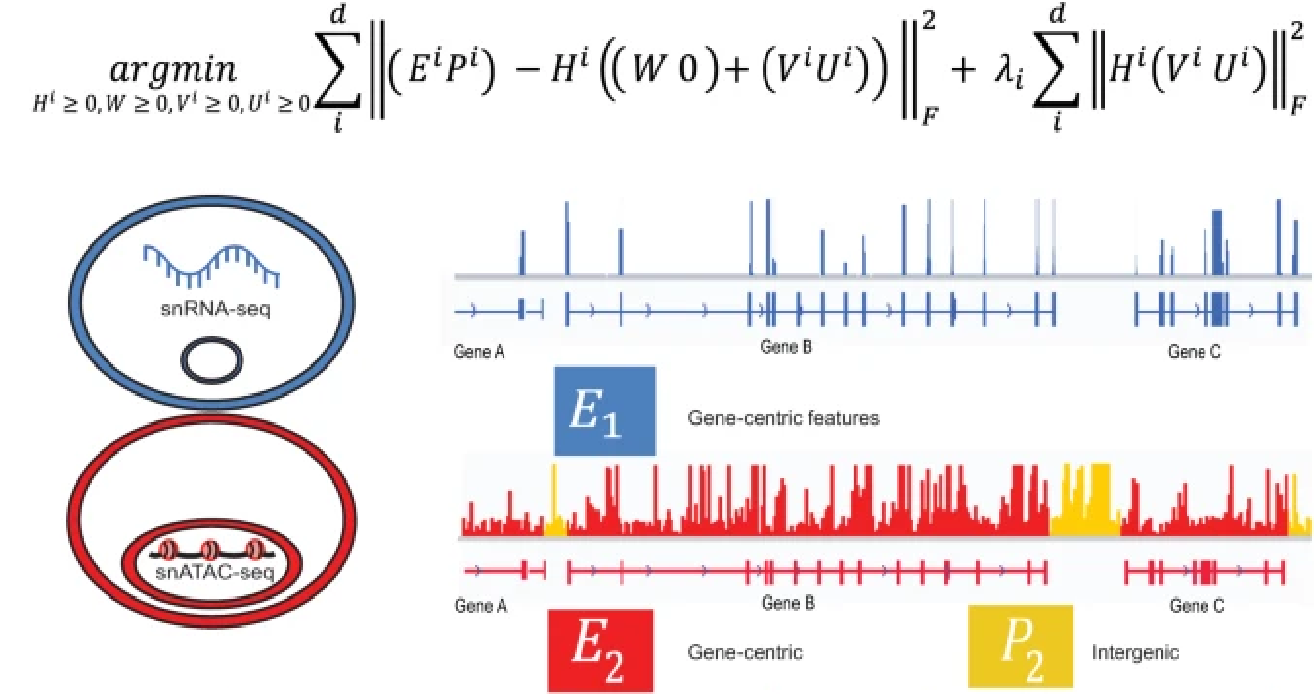
Integration analyses often involve datasets with partially overlapping features, including both shared features that occur in all datasets and features exclusive to a single experiment. To address this limitation, we derive a novel nonnegative matrix factorization algorithm for integrating single-cell datasets containing both shared and unshared features.
Kriebel AR, Welch JD
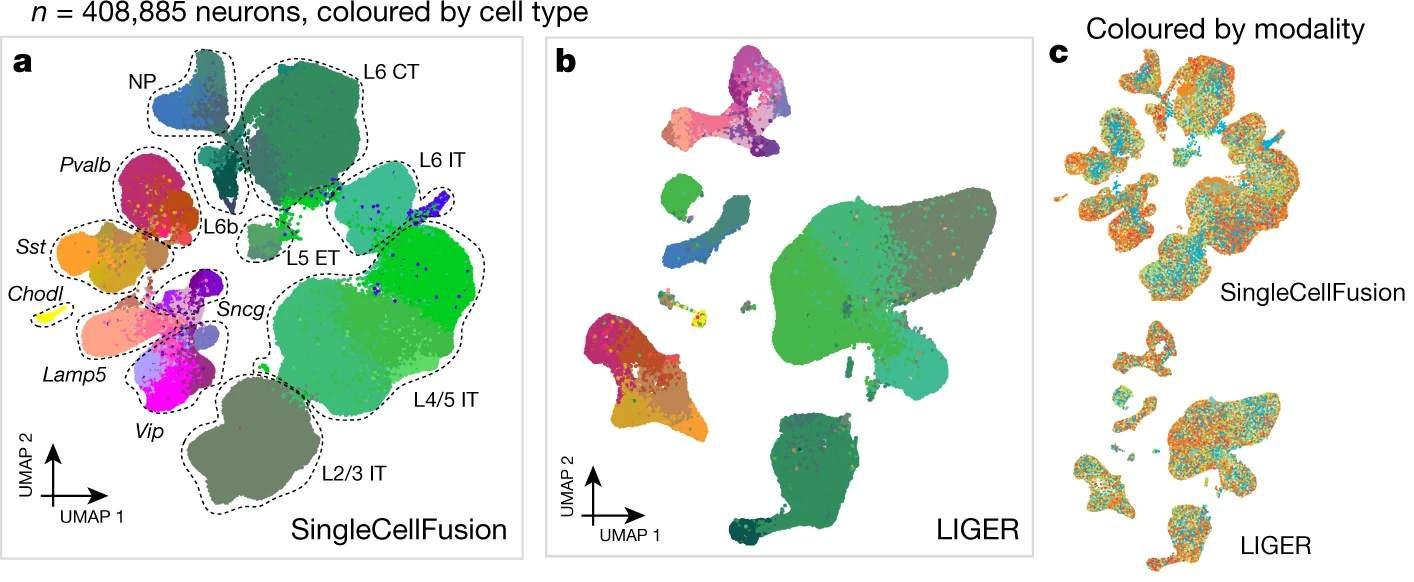
The authors use SingleCellFusion and LIGER to assemble scRNA-seq, snRNA-seq, snmC-seq, and snATAC-seq into an integrated atlas of the diverse cell types in the mouse primary motor cortex.
Zizhen Yao, Hanqing Liu, Fangming Xie, Stephan Fischer, ..., Bosiljka Tasic, Joshua D. Welch, Joseph R. Ecker, Evan Z Macosko, Bing Ren, BRAIN Initiative Cell Census Network (BICCN), Hongkui Zeng, Eran A. Mukamel.
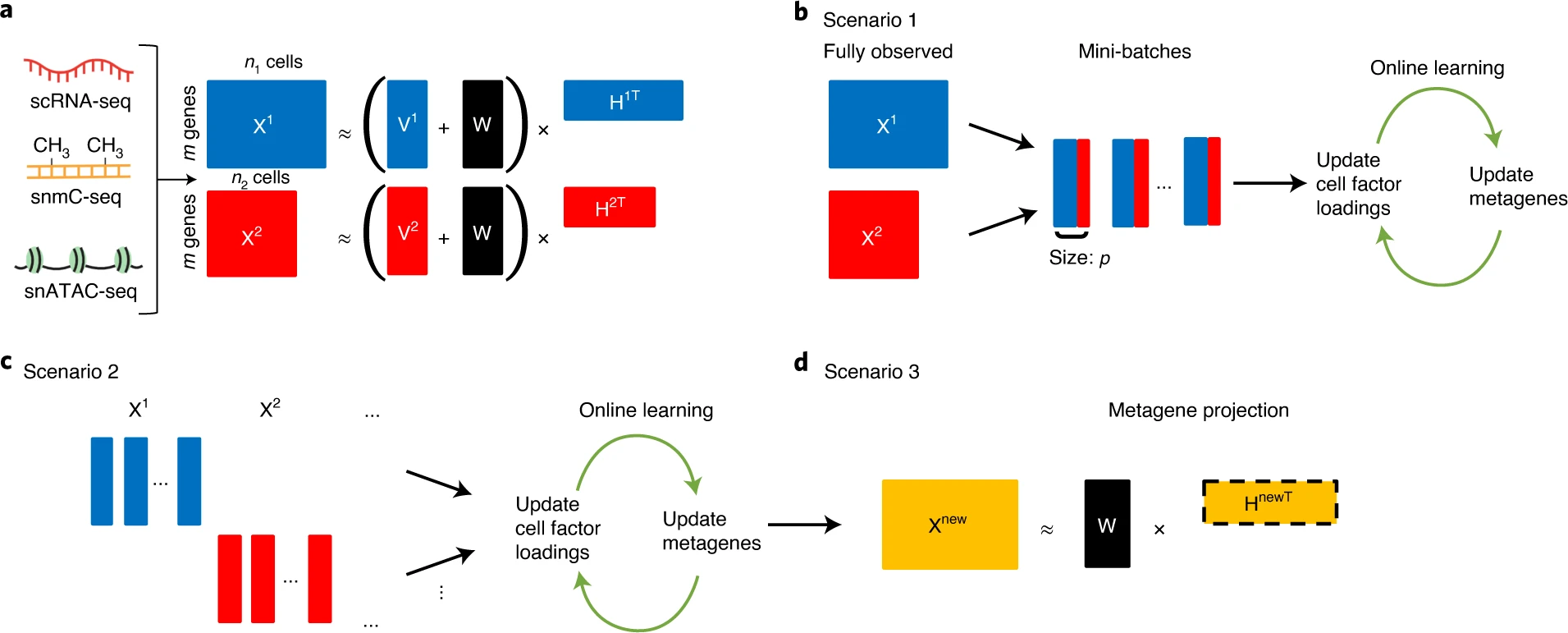
Here we describe online integrative non-negative matrix factorization (iNMF), an algorithm for integrating large, diverse and continually arriving single-cell datasets. Our approach scales to arbitrarily large numbers of cells using fixed memory, iteratively incorporates new datasets as they are generated and allows many users to simultaneously analyze a single copy of a large dataset by streaming it over the internet. Iterative data addition can also be used to map new data to a reference dataset.
Gao C, Liu J, Kriebel AR, Preissl S, Luo C, Castanon R, Sandoval J, Rivkin A, Nery JR, Behrens MM, Ecker JR, Ren B, Welch JD
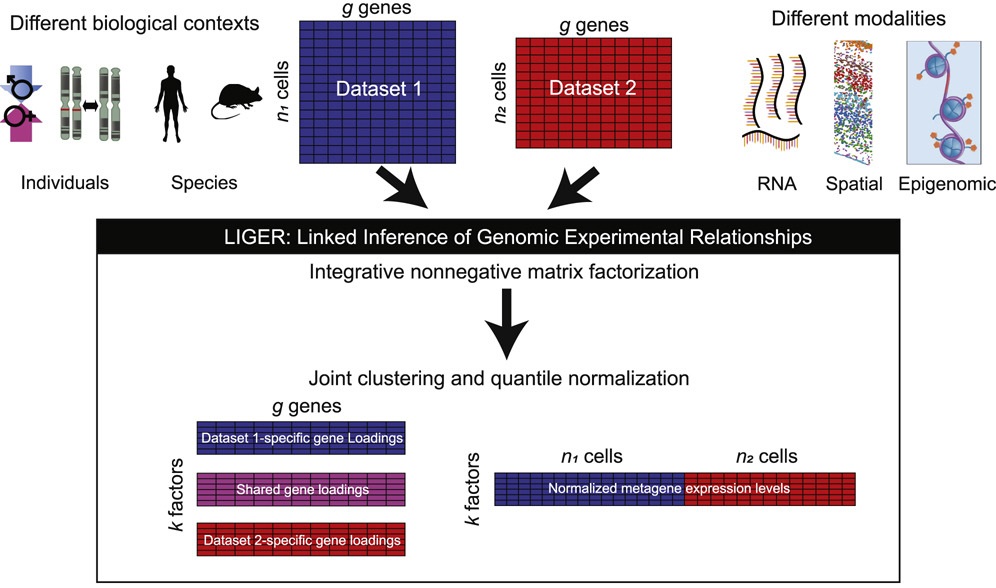
To flexibly model single-cell datasets, we developed LIGER, an algorithm that delineates shared and dataset-specific features of cell identity. We applied it to four diverse and challenging analyses of human and mouse brain cells. Integrative analyses using LIGER promise to accelerate investigations of cell-type definition, gene regulation, and disease states.
Welch JD*, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko EZ*
*Co-Senior Authors
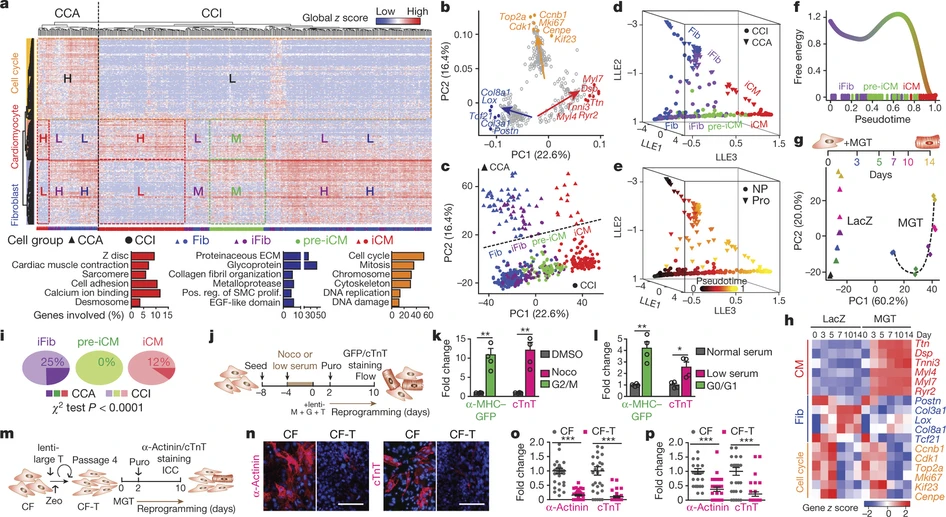
We used single-cell RNA sequencing to analyse global transcriptome changes at early stages during the reprogramming of mouse fibroblasts into induced cardiomyocytes (iCMs). Using unsupervised dimensionality reduction and clustering algorithms, we identified molecularly distinct subpopulations of cells during reprogramming. We also constructed routes of iCM formation, and delineated the relationship between cell proliferation and iCM induction.
Liu Z*, Wang L*, Welch JD*, Ma H, Zhou Y, Vaseghi HR, Yu S, Wall JB, Alimohamadi S, Zheng M, Yin C, Shen W, Prins JF, Liu J, Qian L
*Equal contribution
Full List of publications
Molecular effects of transposable element sequences in mammalian cells
Wen MC, Welch JD
Genome Biology 26, 403 (2025).
Inferring differential dynamics from multi-lineage, multi-omic, and multi-sample single-cell data with MultiVeloVAE
Li C*, Gu Y*, Virgilio MC, Lee KH, Collins KL, Welch JD
Nature Communications 2025
VISTA-FISH: Video Imaging, Spatial Transcriptomics, and CRISPR Screening of the Same Cells
Lee KH*, Yao M*, Ghaemmaghami J*, DeLong CJ, Zhao N, Moline J, Sexton J, Barmada SJ+, Welch JD+
bioRxiv 2025
Predicting the structural impact of human alternative splicing
Song Y, Zhang C, Omenn GS, O’Meara MJ*, Welch JD*
Genome Biology 2025
Mapping cell fate transition in space and time
Gu Y, Welch JD
Nature Biotechnology 2025
Topological velocity inference from spatial transcriptomic data
Gu Y*, Liu J*, Lee KH*, Li C, Lu L, Moline J, Guan R, Welch JD
Nature Biotechnology 2025
PerturbNet predicts single-cell responses to unseen chemical and genetic perturbations
Yu H*, Qian W*, Song Y, Welch JD
Molecular Systems Biology 2025
Wnt-directed CXCL12-expressing apical papilla progenitor cells drive tooth root formation
Nagata, Gadhvi GT, Komori T, Arai Y, Manabe H, Ka Yan Chu A, Kaur R, Ali M, Yang Y, Tsutsumi-Arai C, Nakai Y, Matsushita Y, Tokavanich N, Zheng WJ, Welch JD, Ono N, Ono W.
Nature Communications 16:5510 2025
A Hedgehog–Foxf axis coordinates dental follicle-derived alveolar bone formation
Nagata M, Gadhvi GT, Komori T, Arai Y, Tsutsumi-Arai C, Ka Yan Chu A, Nye SN, Yang Y, Orikasa S, Takahashi A, Carlsson P, Zheng WJ, Welch JD, Ono N, Ono W
Nature Communications 16:6061 2025
CytoSimplex: Visualizing Single-cell Fates and Transitions on a Simplex
Liu J*, Wang Y*, Li C, Gu Y, Ono N, Welch JD
Bioinformatics 2025
Lung CD4+ resident memory T cells use airway secretory cells to stimulate and regulate onset of allergic airway neutrophilic disease
Ravi VR, Korkmaz FT, De Ana CL, Lu L, Shao FZ, Odom CV, Barker KA, Ramanujan A, Niszczak EN, Goltry WN, Martin IM, Ha CT, Quinton LJ, Jones MR, Fine A, Welch JD, Chen F, Belkina AC, Mizgerd JP, Shenoy AT
Cell Reports 2025
Integrating single-cell multimodal epigenomic data using 1D-convolutional neural networks
Gao C, Welch JD
Bioinformatics 2025
Opportunities and challenges of single-cell and spatially resolved genomics methods for neuroscience discovery
Bonev B, Gonçalo CB, Chen F, Codeluppi S, Corces MR, Fan J, Heiman M, Harris K, Inoue F, Kellis M, Levine A, Lotfollahi M, Luo C, Maynard KR, Nitzan M, Ramani V, Satijia R, Schirmer L, Shen Y, Sun N, Green GS, Theis F, Wang X, Welch JD, Gokce O, Konopka G, Liddelow S, Macosko E, Bayraktar O, Habib N, Nowakowski TJ
Nature Neuroscience 2024
Single-cell multi-omic analysis of post-transplant mesenchymal cells reveals molecular signatures and putative regulators of lung fibrosis
Lu L, McLinden AP, Walker NM, Vittal R, Wang Y, Combs MP, Welch JD*, Lama VN*
bioRxiv 2024
TET2 promotes tumor antigen presentation and T cell IFN-γ, which is enhanced by vitamin C
Cheng M, Ka Yan Chu A, Li Z, Yang S, Smith MD, Zhang Q, Brown NG, Marzluff WF, Bardeesy N, Milner JJ, Welch JD, Xiong Y, Baldwin AS
JCI Insight 2024
LUMIC: Latent diffUsion for Multiplexed Images of Cells
Hung AZ, Zhang CJ, Sexton JZ, O'Meara MJ*, Welch JD*
bioRxiv (2024)
Linking transcriptome and morphology in bone cells at cellular resolution with generative AI
Lu L, Ono N*, Welch JD*
Journal of Bone and Mineral Research, zjae151, https://doi.org/10.1093/jbmr/zjae151 (2024)
Deciphering the impact of genomic variation on function
IGVF Consortium
Nature 633, 47-57 (2024)
HIV-1 Vpr combats the PU.1-driven antiviral response in primary human macrophages
Virgilio MC, Ramnani B, Chen T, Disbennett WM, Lubow J, Welch JD, Collins KL
Nature Communications volume 15, Article number: 5514 (2024)
How has the AI boom impacted algorithmic biology?
Singh M, Sahinalp C, Zeng J, Li WV, Kingsford C, Zhang Q, Przytycka T, Welch J, Ma J, Berger B
Cell Systems, Volume 15, Issue 6, P483-487, June 19, 2024
Autophagy and machine learning: Unanswered questions
Yang Y, Pan Z, Sun J, Welch J, Klionsky D.
Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease, Volume 1870, Issue 6, 2024
CytoSignal Detects Locations and Dynamics of Ligand-Receptor Signaling at Cellular Resolution from Spatial Transcriptomic Data
Liu J, Manabe H, Qian W, Wang Y, Gu Y, Ka Yan Chu A, Gadhvi G, Song Y, Ono N, Welch JD
bioRxiv 2024
Endosteal stem cells at the bone-blood interface: A double-edged sword for rapid bone formation
Matsushita Y, Liu J, Ka Yan Chu A, Ono W, Welch JD, Ono N
BioEssays 2023
Predicting the Structural Impact of Human Alternative Splicing
Song Y, Zhang C, Omenn GS, O'Meara MJ*, Welch JD*
bioRxiv 2023
Multiplying insights from perturbation experiments: predicting new perturbation combinations
Welch JD
Molecular Systems Biology 2023
Bone marrow endosteal stem cells dictate active osteogenesis and aggressive tumorigenesis
Matsushita Y*, Liu J*, Ka Yan Chu A, Tsutsumi-Arai C, Nagata M, Arai Y, Ono W, Yamamoto K, Saunders TL, Welch JD*, Ono N*
Nature Communications 2023
Multi-omic single-cell velocity models epigenome–transcriptome interactions and improves cell fate prediction
Li C, Virgilio MC, Collins KL, Welch JD
Nature Biotechnology 41, 387–398 (2023).
MorphNet predicts cell morphology from single-cell gene expression
Lee H, Welch JD
bioRxiv 2022
A bilevel optimization method for tensor recovery under metric learning constraints
Bagherian M, Tarzanagh DA, Dinov I, Welch JD
arXiv 2022
Bayesian inference of RNA velocity from multi-lineage single-cell data
Gu Y, Blaauw D, Welch JD
bioRxiv 2022
Variational mixtures of ODEs for inferring cellular gene expression dynamics
Gu Y, Blaauw D, Welch JD
ICML 2022
UINMF performs mosaic integration of single-cell multi-omic datasets using nonnegative matrix factorization
Kriebel AR, Welch JD
Nature Communications 13:780 (2022)
PyLiger: Scalable single-cell multi-omic data integration in Python
Lu L, Welch JD
Bioinformatics, btac190, March 2022
SquiggleNet: Real-Time, Direct Classification of Nanopore Signals
Bao Y, Welch JD
Genome Biology 22\:298 (2021)
A multimodal cell census and atlas of the mammalian primary motor cortex
BRAIN Initiative Cell Census Network (BICCN)
Nature 598, 86–102 (2021)
A transcriptomic and epigenomic cell atlas of the mouse primary motor cortex
Zizhen Yao, Hanqing Liu, Fangming Xie, Stephan Fischer, ..., Bosiljka Tasic, Joshua D. Welch, Joseph R. Ecker, Evan Z Macosko, Bing Ren, BRAIN Initiative Cell Census Network (BICCN), Hongkui Zeng, Eran A. Mukamel.
Nature 598, 103–110 (2021)
Functional coordination of non-myocytes plays a key role in adult zebrafish heart regeneration
Ma H, Liu Z, Yang Y, Feng D, Dong Y, Garbutt T, Hu Z, Wang L, Luan C, Cooper C, Li Y, Welch JD*, Qian L*, Liu J*
EMBO Rep (2021) e52901
G-CSF secreted by mutant IDH1 glioma stem cells abolishes myeloid cell immunosuppression and enhances the efficacy of immunotherapy
Alghamri MS, Avvari RP, Thalla R, Kamran N, Zhang L, Ventosa M, Taher A, Faisal SM, Núñez FJ, Fabiani MBG, Haase S, Carney S, Orringer D, Hervey-Jumper S, Heth J, Patil PG, Al-Holou WN, Eddy K, Merajver S, Ulintz PJ, Welch JD, Gao C, Liu J, Núñez G, Hambardzumyan D, Lowenstein PR, Castro M
Science Advances Vol 7, Issue 40 (2021)
Transcription factor FOXF1 identifies compartmentally distinct mesenchymal cells with a role in lung allograft fibrogenesis
Braeuer RR, Walker NM, Misumi K, Mazzoni-Putman S, Aoki Y, Liao R, Vittal R, Kleer GG, Wheeler DS, Sexton JZ, Farver CF, Welch JD, Lama VN
Journal of Clinical Investigation. doi:10.1172/JCI147343
Community-wide hackathons to identify central themes in single-cell multi-omics
Lê Cao KA, Abadi AJ, Davis-Marcisak EF, Hsu L, Arora A, Coullomb A, Deshpande A, Feng Y, Jeganathan P, Loth M, Meng C, Mu W, Pancaldi V, Sankaran K, Singh A, Sodicoff JS, Stein-O’Brien GL, Subramanian A, Welch JD, You Y, Argelaguet R, Carey VJ, Dries R, Greene CS, Holmes S, Love MI, Ritchie ME, Yuan GC, Culhane AC, Fertig E
Genome Biology 22, 220 (2021)
Single-cell transcriptomic analysis reveals developmental relationships and specific markers of mouse periodontium cellular subsets. Frontiers in Dental Medicine
Nagata M, Ka Yan Chu A, Ono N, Welch JD, Ono W
Frontiers in Dental Medicine 2\:56 (2021)
Intercellular interactions of an adipogenic CXCL12‐expressing stromal cell subset in murine bone marrow
Matsushita Y, Ka Yan Chu A, Ono W, Welch JD, Ono N
Journal of Bone and Mineral Research 2021
MichiGAN: Sampling from disentangled representations of single-cell data using generative adversarial networks
Yu H, Welch JD
Genome Biology 22, 158 (2021)
Iterative Single-Cell Multi-Omic Integration Using Online Learning
Gao C, Liu J, Kriebel AR, Preissl S, Luo C, Castanon R, Sandoval J, Rivkin A, Nery JR, Behrens MM, Ecker JR, Ren B, Welch JD
Nature Biotechnology 39, 1000–1007 (2021)
Jointly defining cell types from multiple single-cell datasets using LIGER
Liu J*, Gao C*, Sodicoff J, Kozareva V, Macosko EZ, Welch JD
Nature Protocols (2020)
A Wnt-mediated transformation of the bone marrow stromal cell identity orchestrates skeletal regeneration
Matsushita Y, Nagata M, Kozloff K, Welch J, Mizuhashi K, Tokavanich N, Hallett S, Link D, Nagasawa T, Ono W, and Ono N
Nature Communications. 11, 332 (2020)
Single-Cell Transcriptomic Analyses of Cell Fate Transitions during Human Cardiac Reprogramming
Zhou Y, Liu Z, Welch JD, Gao X, Wang L, Garbutt T, Keepers B, Ma H, Prins JF, Shen W, Liu J, Qian L
Stem Cell. 2019 Jun 12
Single-cell multi-omic integration compares and contrasts features of brain cell identity
Welch JD*, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko EZ*
Cell. Volume 177, Issue 7, 13 June 2019, Pages 1873-1887.e17
Slide seq: A Scalable Technology for Measuring Genome-Wide Expression at High Spatial Resolution
Rodriques SG*, Stickels RR*, Goeva A Martin CA, Murray E, Vanderburg CR, Welch JD, Chen LM, Chen F+, Macosko EZ+
Science 29 Mar 2019: Vol. 363, Issue 6434, pp. 1463-1467
Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte
Liu Z*, Wang L*, Welch JD*, Ma H, Zhou Y, Vaseghi HR, Yu S, Wall JB, Alimohamadi S, Zheng M, Yin C, Shen W, Prins JF, Liu J, Qian L
Nature (02 November 2017). 551, 100–104 doi:10.1038/nature24454
MATCHER: manifold alignment reveals correspondence between single cell transcriptome and epigenome dynamics
Welch JD, Hartemink AJ, Prins JF
Genome Biology 2017.
Selective Single Cell Isolation for Genomics Using Microraft Arrays
Welch JD*, Williams LA*, DiSalvo M*, Brandt AT, Marayati R, Sims CE, Allbritton NL, Prins JF, Yeh JJ, Jones CD
Nucleic Acids Research 2016
A subset of replication-dependent histone mRNAs are expressed as polyadenylated RNAs in terminally differentiated tissues
Lyons SM, Cunningham CH, Welch JD, Groh B, Guo AY, Wei B, Whitfield ML, Xiong Y, Marzluff WF
Nucleic Acids Research 2016
SLICER: Inferring branched, nonlinear cellular trajectories from single cell RNA-seq data
Welch JD, Hartemink AJ, Prins JF
Genome Biology 2016, 17
Robust Detection of Alternative Splicing in a Population of Single Cells
Welch JD, Hu Y, Prins JF
Nucleic Acids Research 2016
A multi-protein occupancy map of the mRNP on the 3 end of histone mRNAs
Brooks L, Lyons SM, Mahoney JM, Welch JD, Liu Z, Marzluff WM, Whitfield ML
RNA 2015
EnD-Seq and AppEnD: Sequencing 3 ends to identify nontemplated tails and degradation intermediates
Welch JD*, Slevin MK*, Tatomer D, Duronio RJ, Prins JF, Marzluff WF
RNA 2015
Pseudogenes Transcribed in Breast Invasive Carcinoma Show Subtype-Specific Expression and ceRNA Potential
Welch JD, Baran-Gale J, Perou C, Sethupathy P, Prins JF
BMC Genomics 2015
Deep Sequencing Shows Multiple Oligouridylations Are Required for 3′ to 5′ Degradation of Histone mRNAs on Polyribosomes
Slevin MK, Meaux S, Welch JD, Bigler R, Miliani de Marval PL, Su W, Rhoads RE, Prins JF, Marzluff WF
Molecular cell 53 (6), 1020-1030. March 2014
WordSeeker: concurrent bioinformatics software for discovering genome-wide patterns and word-based genomic signatures
Lichtenberg J, Kurz K, Liang X, Al-ouran R, Neiman L, Nau LJ, Welch JD, Jacox E, Bitterman T, Ecker K, Elnitski L, Drews F, Lee SS, Welch LR
BMC Bioinformatics. 2010 Dec 21;11 Suppl 12
The word landscape of the non-coding segments of the Arabidopsis thaliana genome. BMC Genomics
Lichtenberg J, Yilmaz A, Welch JD, Kurz K, Liang X, Drews F, Ecker K, Lee SS, Geisler M, Grotewold E, Welch LR
BMC Genomics. 2009 Oct 8
Word-based characterization of promoters involved in human DNA repair pathways
Lichtenberg J, Jacox E, Welch JD, Kurz K, Liang X, Yang MQ, Drews F, Ecker K, Lee SS, Elnitski L, Welch LR
BMC Genomics. 2009 Jul 7;10 Suppl 1